Research Focus
Current Projects
Listeria monocytogenes is an important agent of food-borne infections and has been responsible for some of the most extensive and expensive food recalls. This bacterium is a ubiquitous environmental bacterium that can survive in the soil as a saprophyte, but which maintains an ability to invade and replicate within the cells of susceptible humans and animals. While environmental microorganisms are numerous, relatively few have the capacity to cause human disease, thus we are interested in how L. monocytogenes balances its life as an environmental organism with the lifestyle of a human/animal pathogen. Our studies take advantage of the fact that L. monocytogenes is very amenable to genetic manipulation, and that excellent cell culture and animal infections models exist to study the molecular mechanisms of bacterial pathogenesis. Current work is focused on defining the role of small bacterial peptide signaling molecules in promoting L. monocytogenes intracellular replication, and on the mechanisms of virulence factor secretion. L. monocytogenes appears to maintain its virulence arsenal by exploiting secreted factors for a variety of activities. For example, a secreted bacterial chitinase that functions to break down chitin in the outside environment also suppresses the expression of host innate immune responses that normally help to limit bacterial infection. We are also interested in determining what contributes to host tissue tropism during bacterial infection. L. monocytogenes has been well associated with infections of the central nervous system and with fetal infections during pregnancy, but we have recently found that some clinical isolates also have the capacity to cause cardiac infections. We are therefore working to define which bacterial factors define where and how L. monocytogenes chooses to replicate within an infected host. Preliminary experiments suggest that the bacterium may manipulate host cell metabolism to improve intracellular bacterial replication. We are also interested in determining how maternal and fetal immune systems contribute to resistance to Listeria infection.
Defining the impact of anesthesia on host susceptibility to microbial infection
Defining the impact of anesthesia on host susceptibility to microbial infection
Additional studies in the lab are exploring how common anesthetic agents increase host susceptibility to microbial infection. Infections acquired following surgical procedures account for billions of dollars in health care costs and have severe negative impact on patient outcomes. We are currently using models of Staphylococcus aureus and Klebsiella pneumoniae infection in addition to L. monocytogenes to decipher how anesthetic agents influence host immune responses and to develop new strategies to limit hospital acquired infections.
In our studies we use propofol, one of the most common anesthetic agents used in surgery and for routine outpatient procedures, such as gastrointestinal endoscopy. It has been previously found in vitro that immune responses, such as secretion of certain inflammatory immunocytokines, can be modulated by propofol. Using in vivo mouse models of infection, we found that propofol significantly enhances host susceptibility to pathogen infection by inhibiting the recruitment and/or activity of immune effector cells at sites of infection. In a separate study, our data indicated that short-term sedation with propofol significantly increases the severity of bloodstream MRSA infection, even in conjunction with vancomycin prophylaxis.
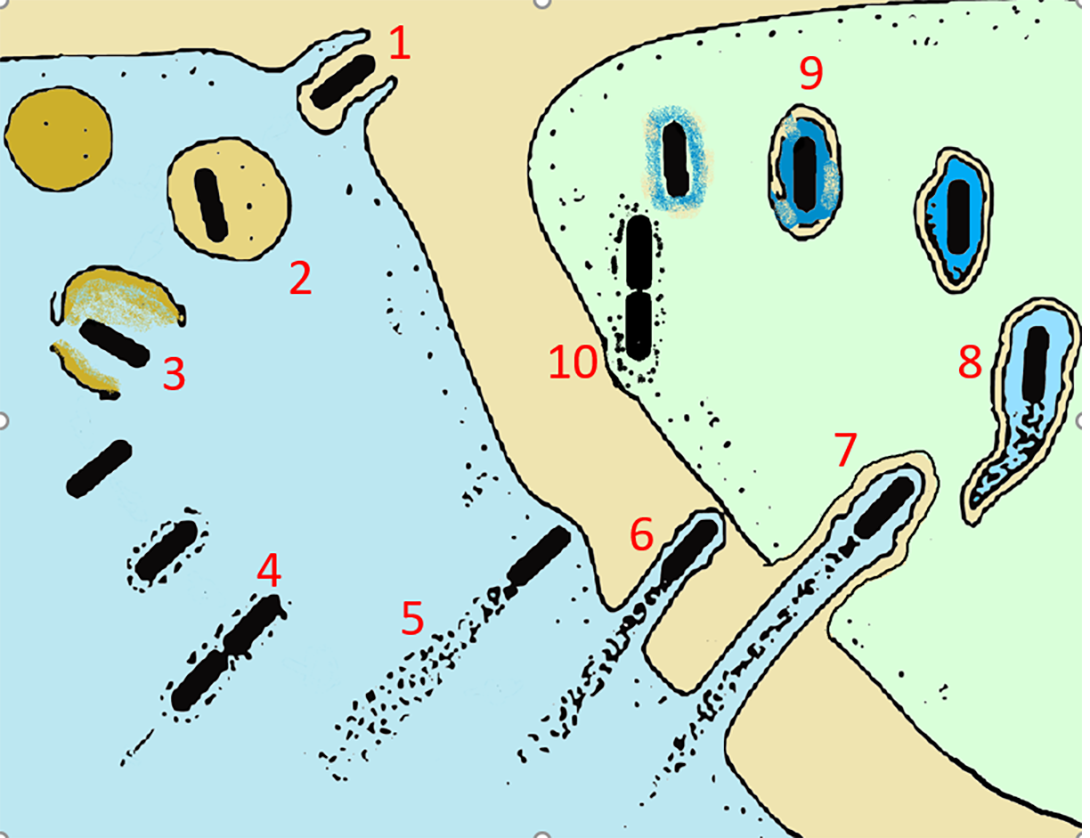
ABOVE RIGHT: 1. Entry into the host cell by induced phagocytosis 2. Transient residence within a phagocytic vacuole 3. Escape from the phagosome into the cytoplasm and PrfA activation 4. Cytosolic replication and recruitment of host cell actin onto the bacterial surface 5. Actin-based motility 6. Formation of pseudopods 7. Phagocytosis of the pseudopods by neighboring cells 8. Formation of a double-membrane phagosome 9. Escape from this secondary phagosome 10. Reinitiation of the cycle
Bacterial infections during pregnancy
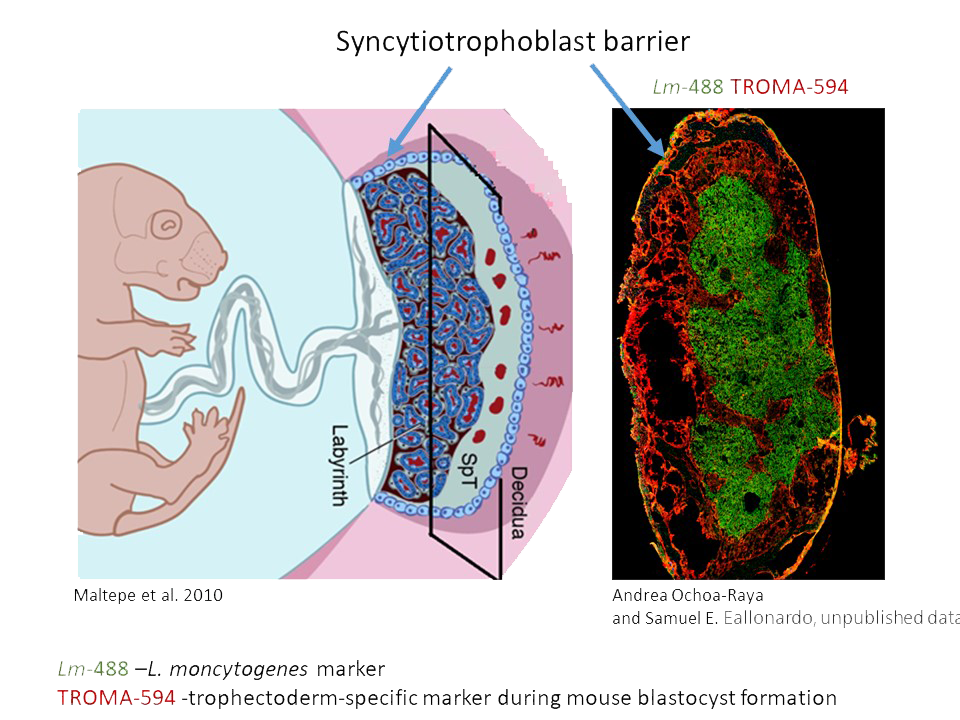
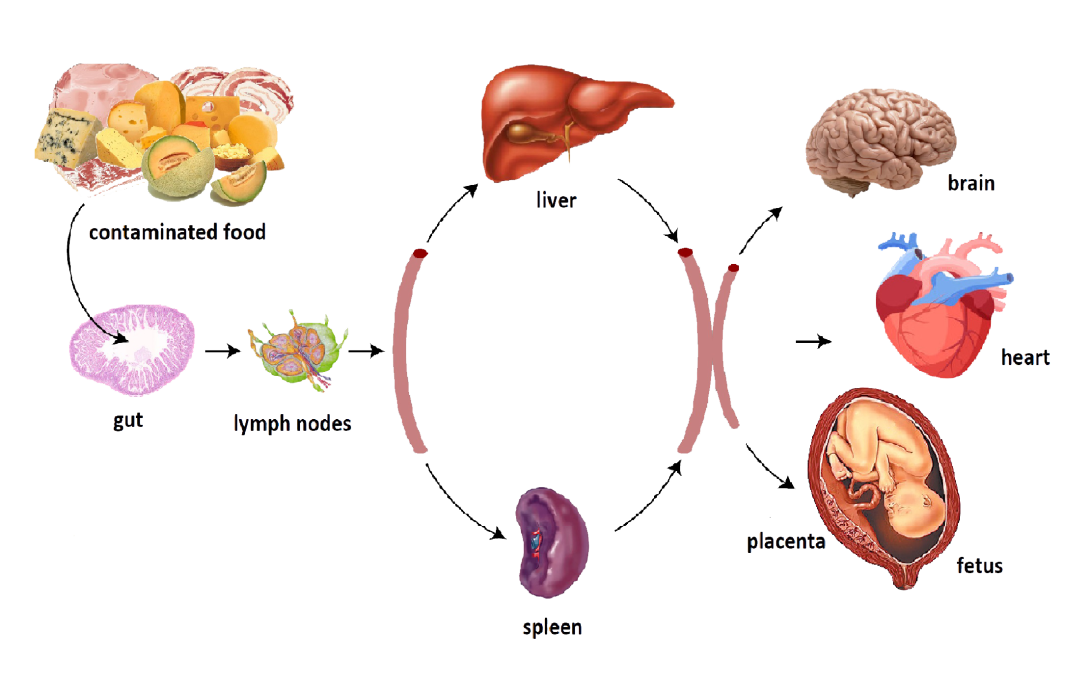
Bacterial infections during pregnancy
Many Listeria monocytogenes infections during pregnancy go unnoticed or appear as mild, non-specific infections of the mother. However, they can result in serious fetal and neonatal consequences to a child, leading to preterm births, miscarriages, and stillbirth. Our studies indicate that clinical strains of Listeria infect placenta most readily when high levels of bacterial surface protein InlB are present. InlB interacts with mammalian c-Met, or hepatocyte growth factor (HGFR) receptor, abundant on the surface of epithelial and endothelial cells of placenta and fetus. C-Met is essential for embryonic development and tissue repair, but unfortunately due to its interaction with bacterial InlB, it also mediates internalization and infection of placenta and fetus with Listeria monocytogenes. Using the clinical strains of Listeria and the pregnant mouse models of infection, we discovered that increased expression of InlB on the surface of Listeria is sufficient to convey high frequency vertical transmission from mother to fetus. Our goal is to characterize the structure and regulation of bacterial InlB and the role of signaling resulting from the interaction of InlB with c-Met in spreading of infection. The innate immune system at the maternal/fetal interface mediates a delicate balance between being robust enough to protect the fetus from maternal pathogens but measured enough to prevent immune rejection of semi-allogenic fetus and placenta by the mother. We are interested in characterizing responses of placental and fetal innate immunity to Lm infections and contributions of MCP-1, TNFα, or IFNγ to fetal immune defenses. In addition, we investigate the tropism, time course, and spatial resolution of Listeria infection in pregnant mouse and human placentas.
CD45+ Cells are Found in the Mouse Placenta
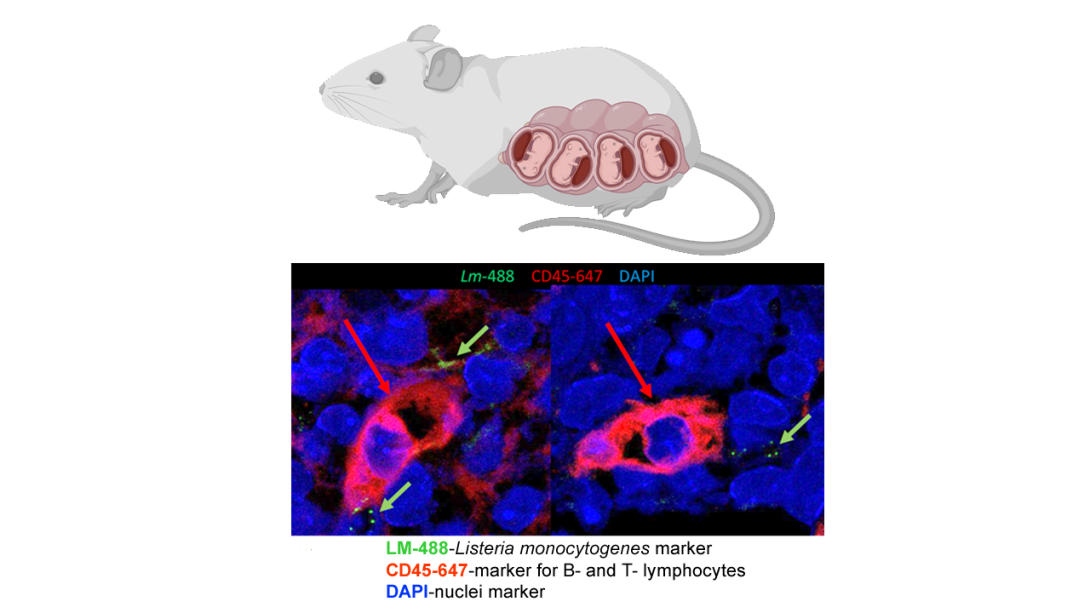
ABOVE: The immunofluorescent image of the slice of mouse placenta from infected mother indicates that Listeria monocytogenes is capable of infecting maternal placenta (green arrows). In addition, one can observe an infiltration of placenta with B and T-lymphocytes (red arrow), suggesting a mounted immune response in the placenta against the invading pathogen.
Regulation of heme transport by peptide phermone pPplA
Regulation of heme transport by peptide phermone pPplA
Upon infection, bacterial pathogens must scavenge all nutrients required for their survival from the host’s body. One such essential micronutrient is iron, which is found complexed to host’s iron-binding proteins such as hemoglobin, ferritin, transferrin and others, and thus not easily accessible to bacteria for their own unique needs.
To overcome the “nutritional immunity” or sequestration of micronutrients by the host, Listeria evolved specific mechanisms for extraction and transport of iron. Three major pathways of iron acquisition by Lm have been described so far. They include reductive transport of ferric iron via FepA/B/C transporter; uptake of iron in the form of ferrichrome and ferrioxamine xenosiderophores by FhuB/C/D/G ATPase transporter; and uptake of iron-rich heme via HupC/D/G transport complex. Based on RNAseq we found the latter system to be negatively regulated at the level of transcription by a small peptide pheromone pPplA, previously discovered in our lab.
We hypothesize that in confined space, such as in a vacuole, the concentraion of pPplA is high and this serves to suppress the iron/heme uptake but stimulates the mechanism of bacterial escape from the vacuole. Once a bacterium breaks free and starts infiltrating the host’s blood vessels, the secreted pPplA peptides diffuse away into the blood stream, essentially abolishing the outside-in pheromone signaling. This event could potentially serve as a cue for Lm to express heme transport genes for uptake of heme, a major source of iron in the blood. Surprisingly, we found that low micromolar concentrations of hemin (Hn), an oxidized form of heme, initially inhibit the L. m growth in the chemically defined Welshimer’s Broth within the first 24 hrs. After that bacteria adapt to heme as the only source of iron and begin to grow at the normal rate. The mechanism responsible for an adaptation of bacteria to heme toxicity, as well as the regulation of heme transport by pPplA, are currently under investigation.